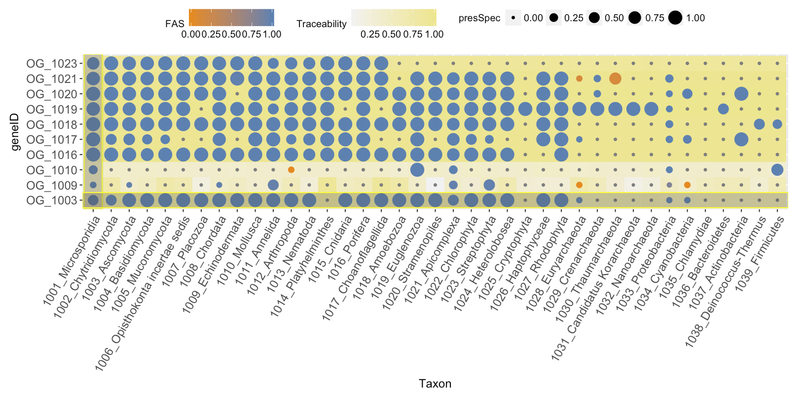
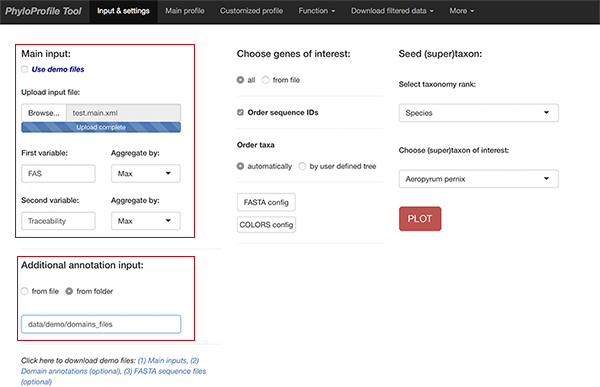
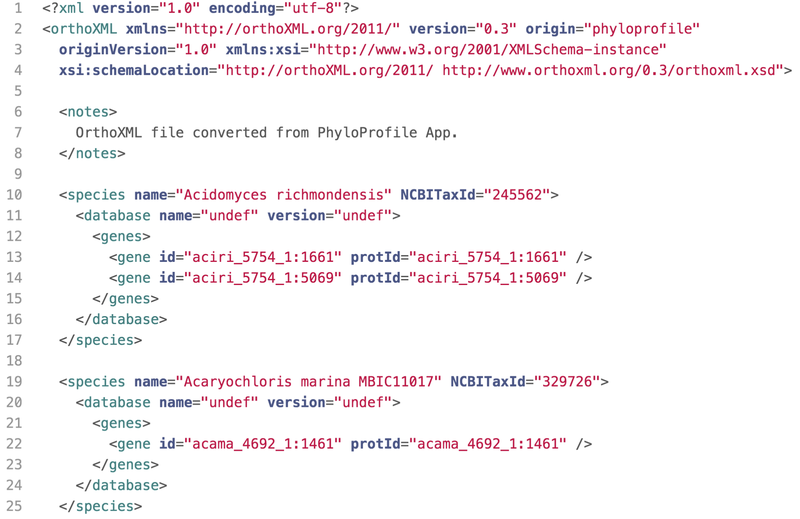
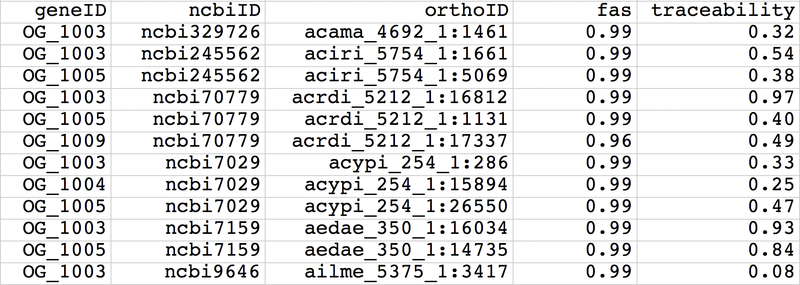
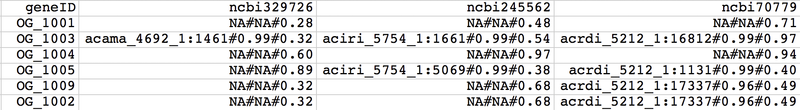
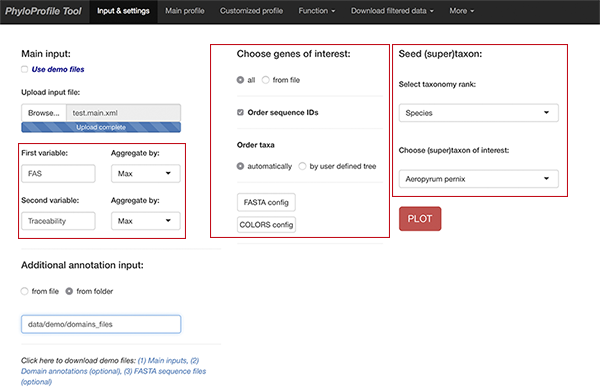
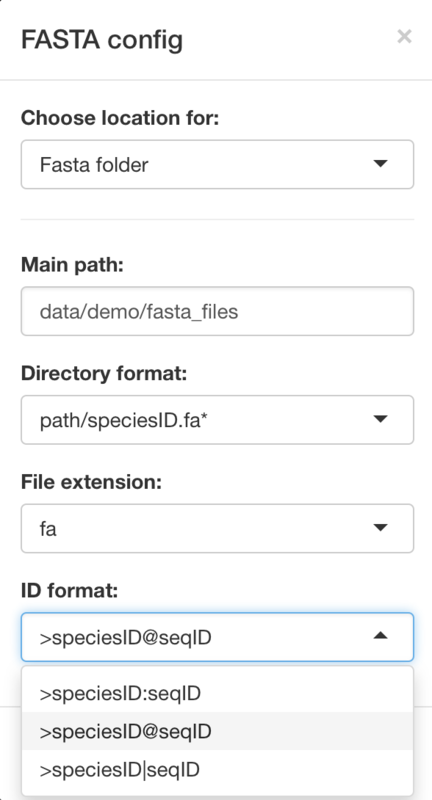
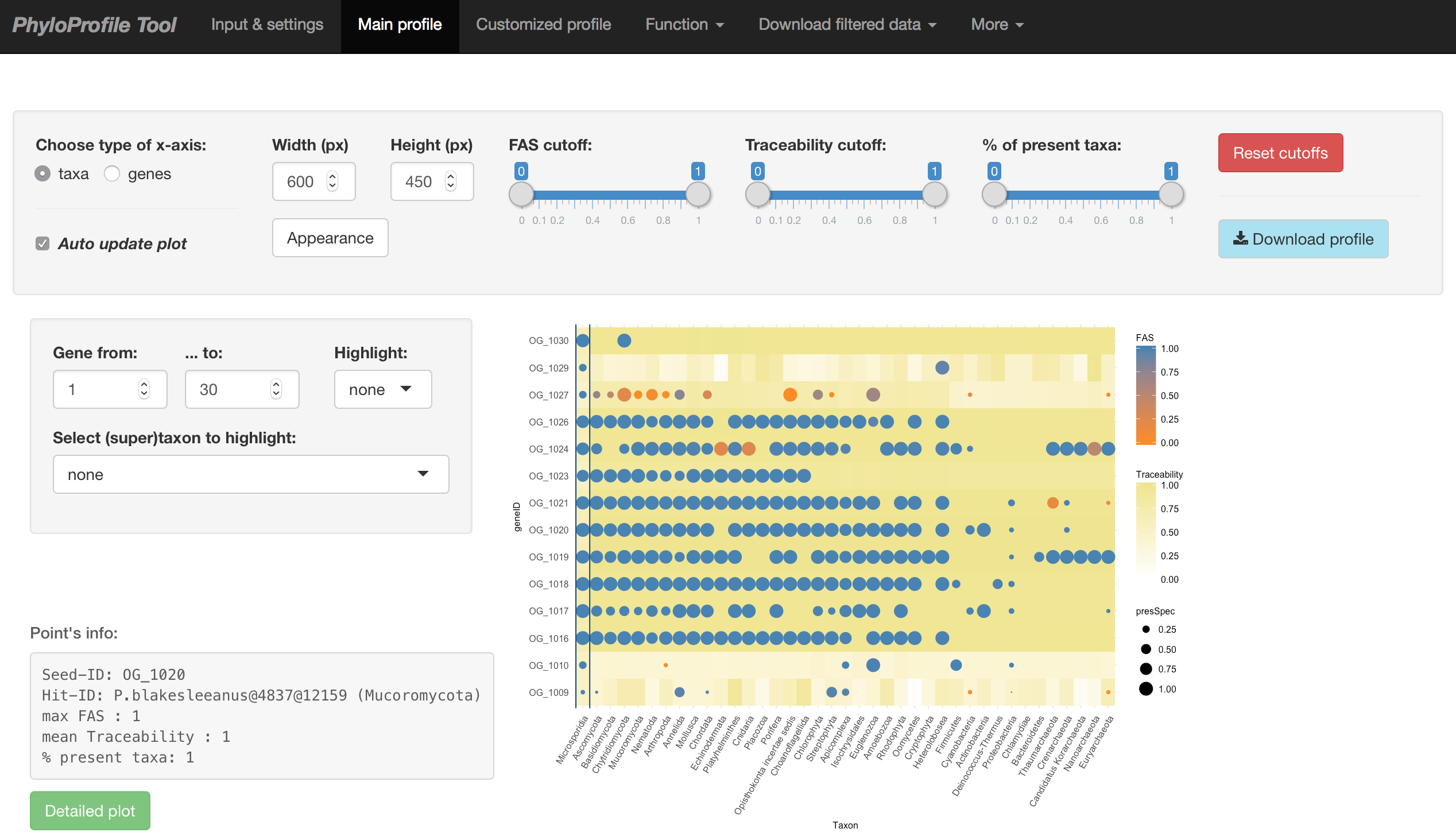
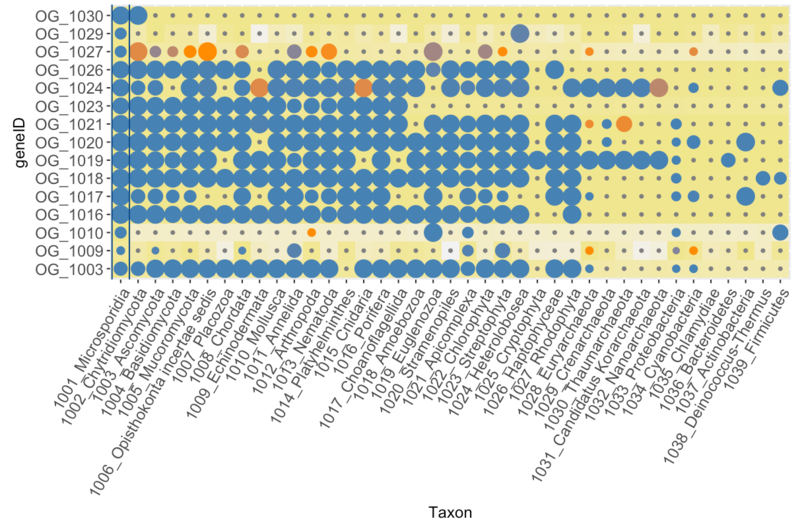
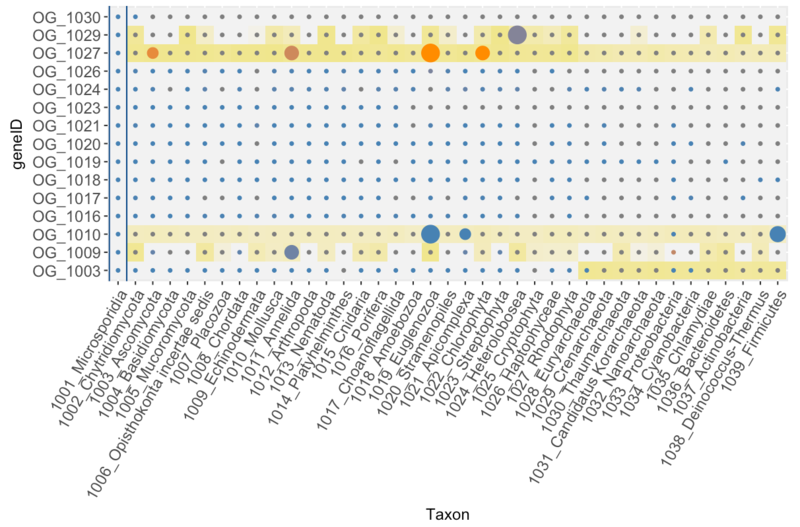
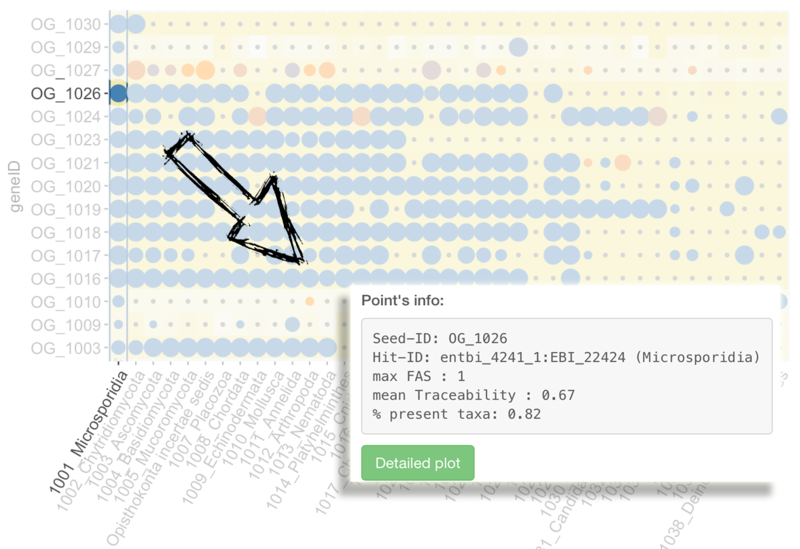
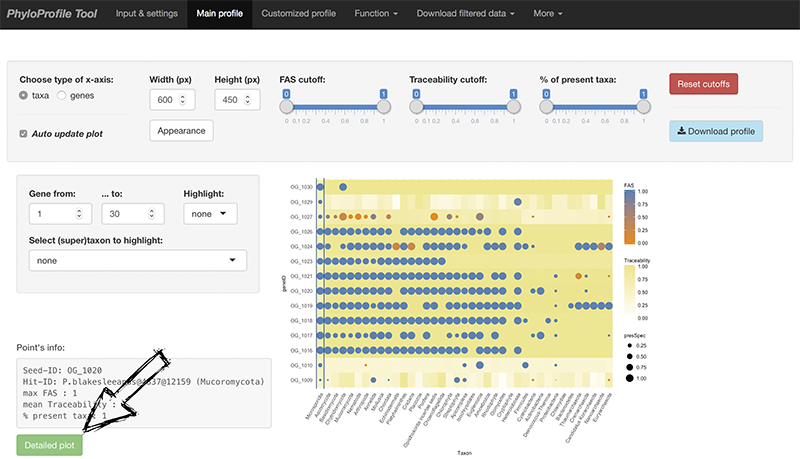
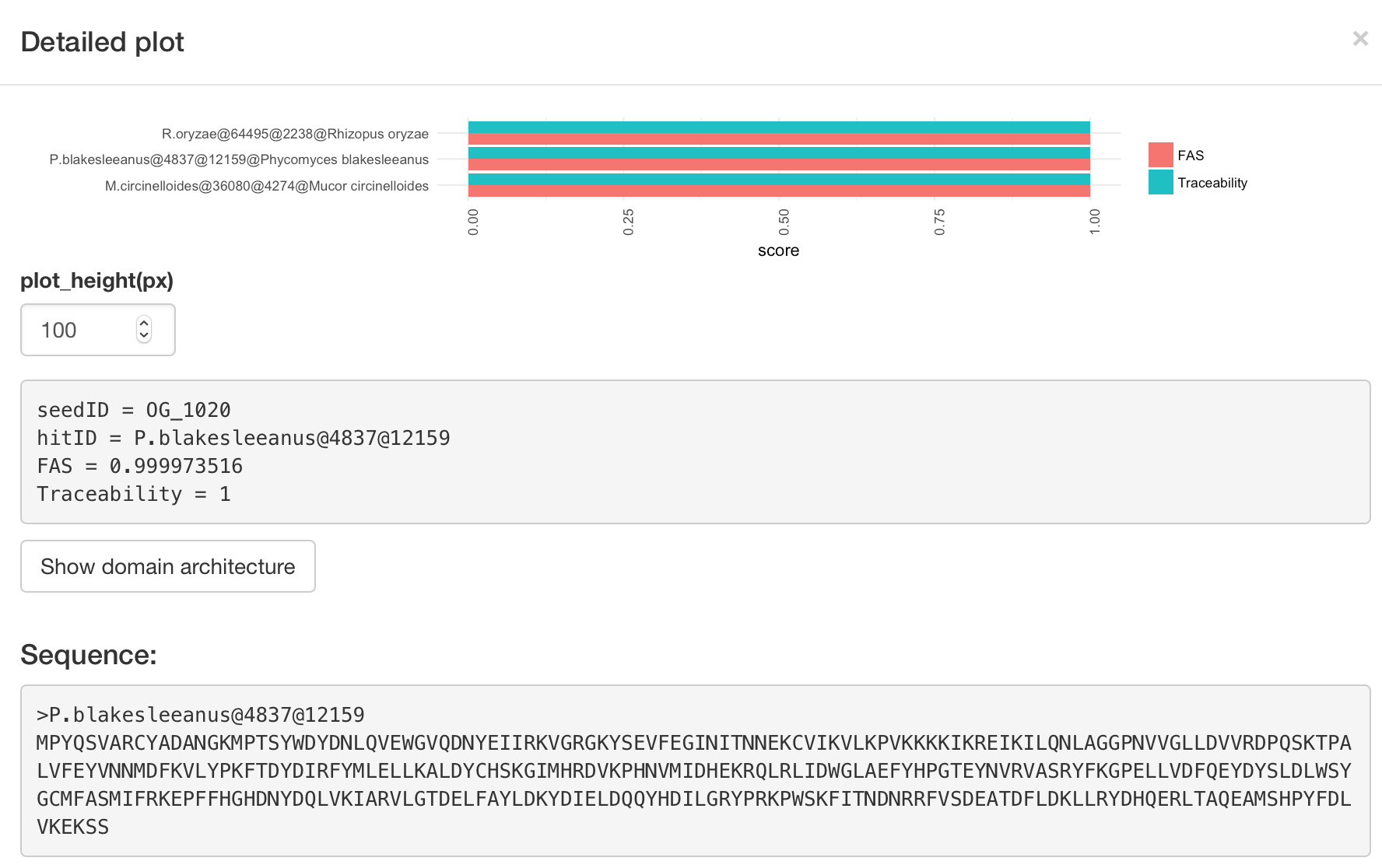
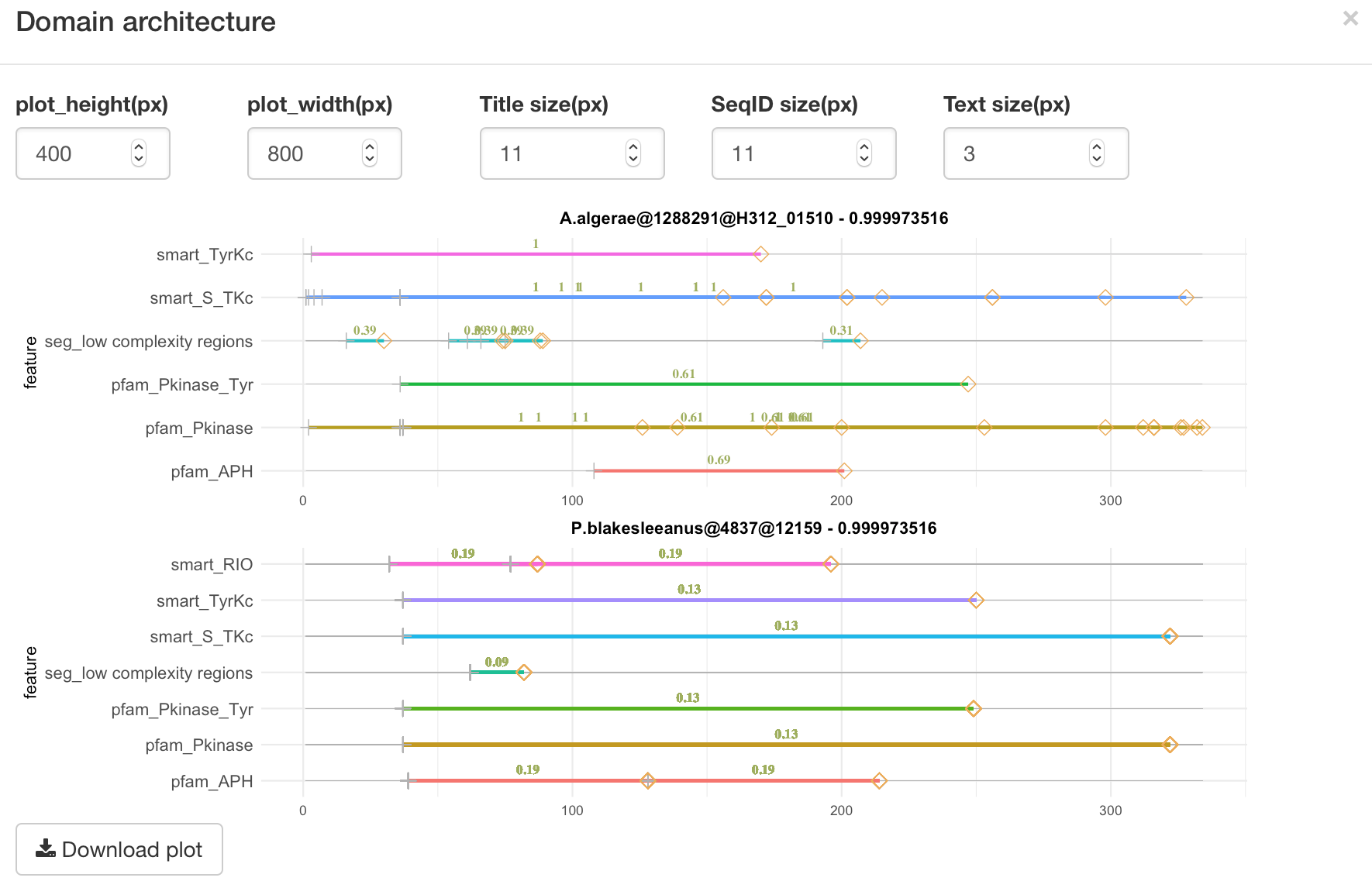
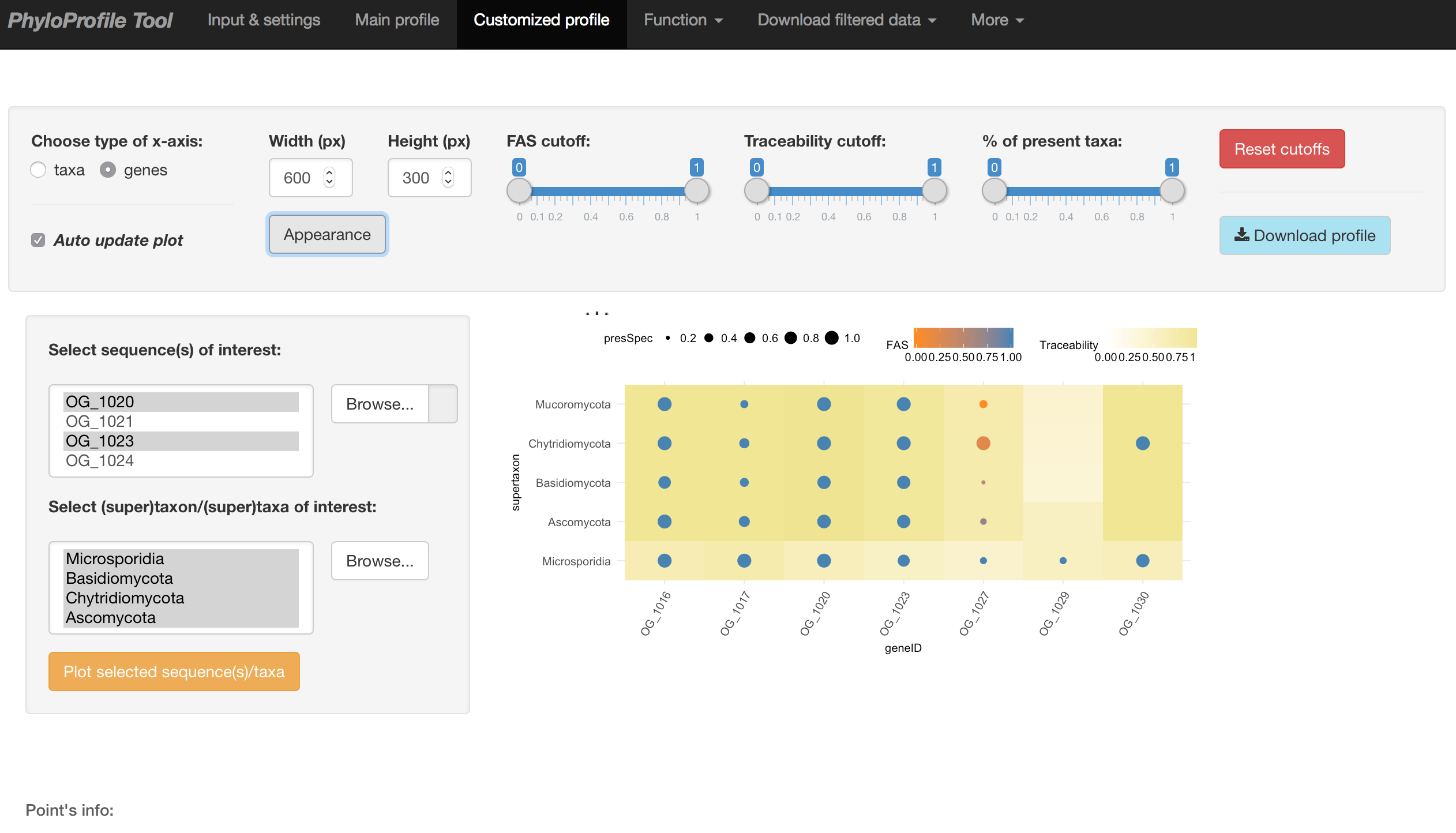
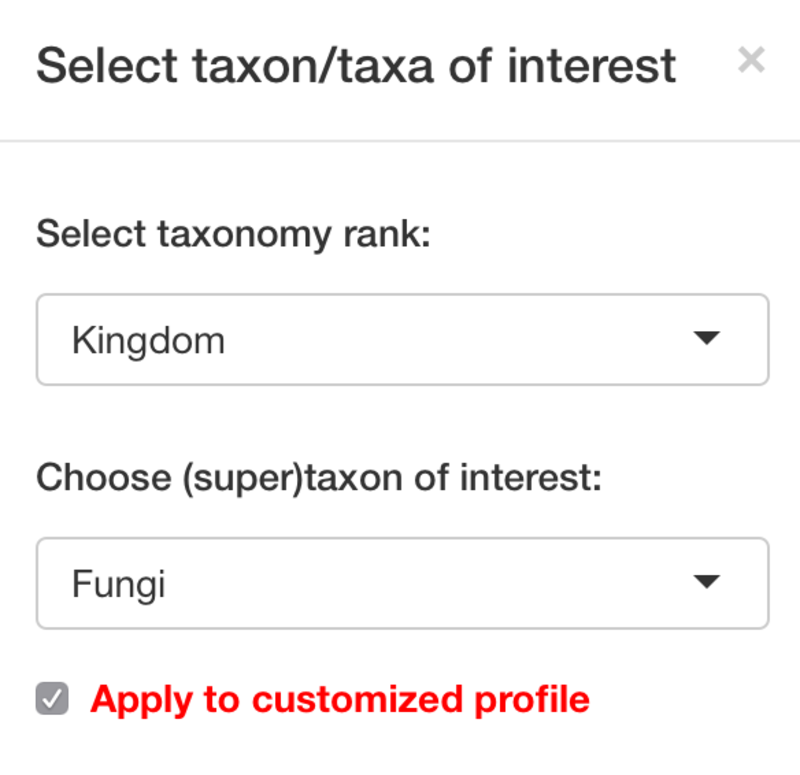
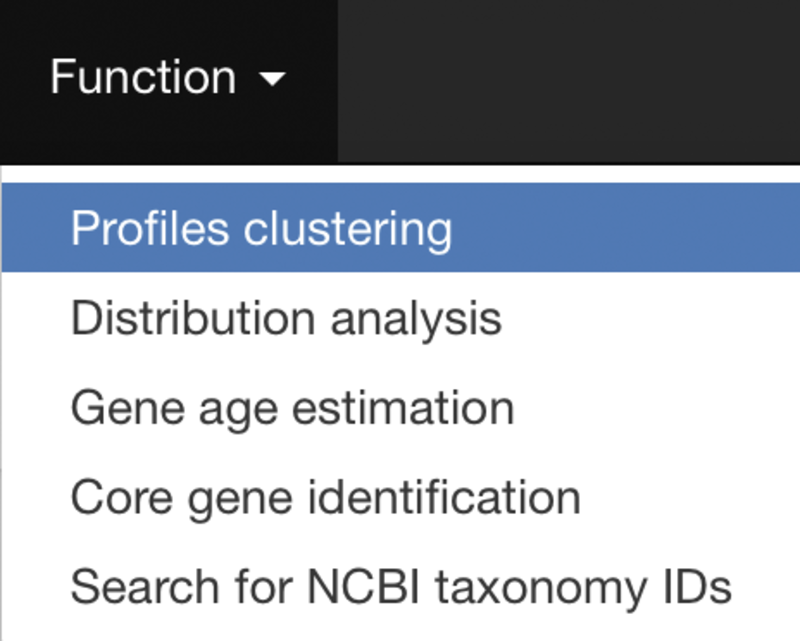
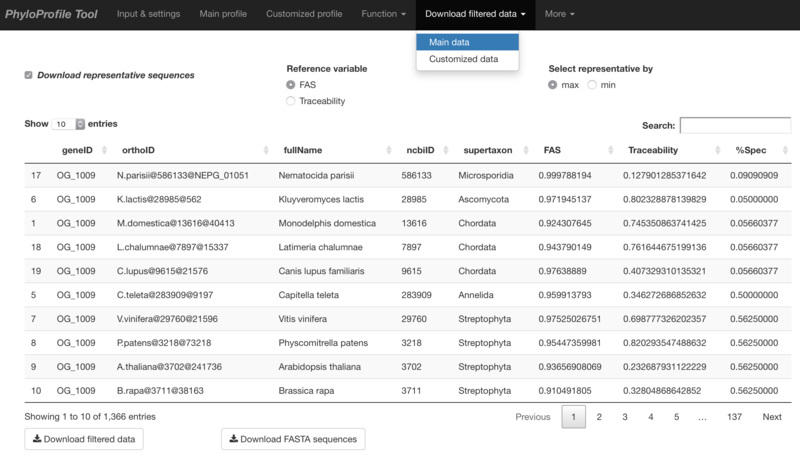
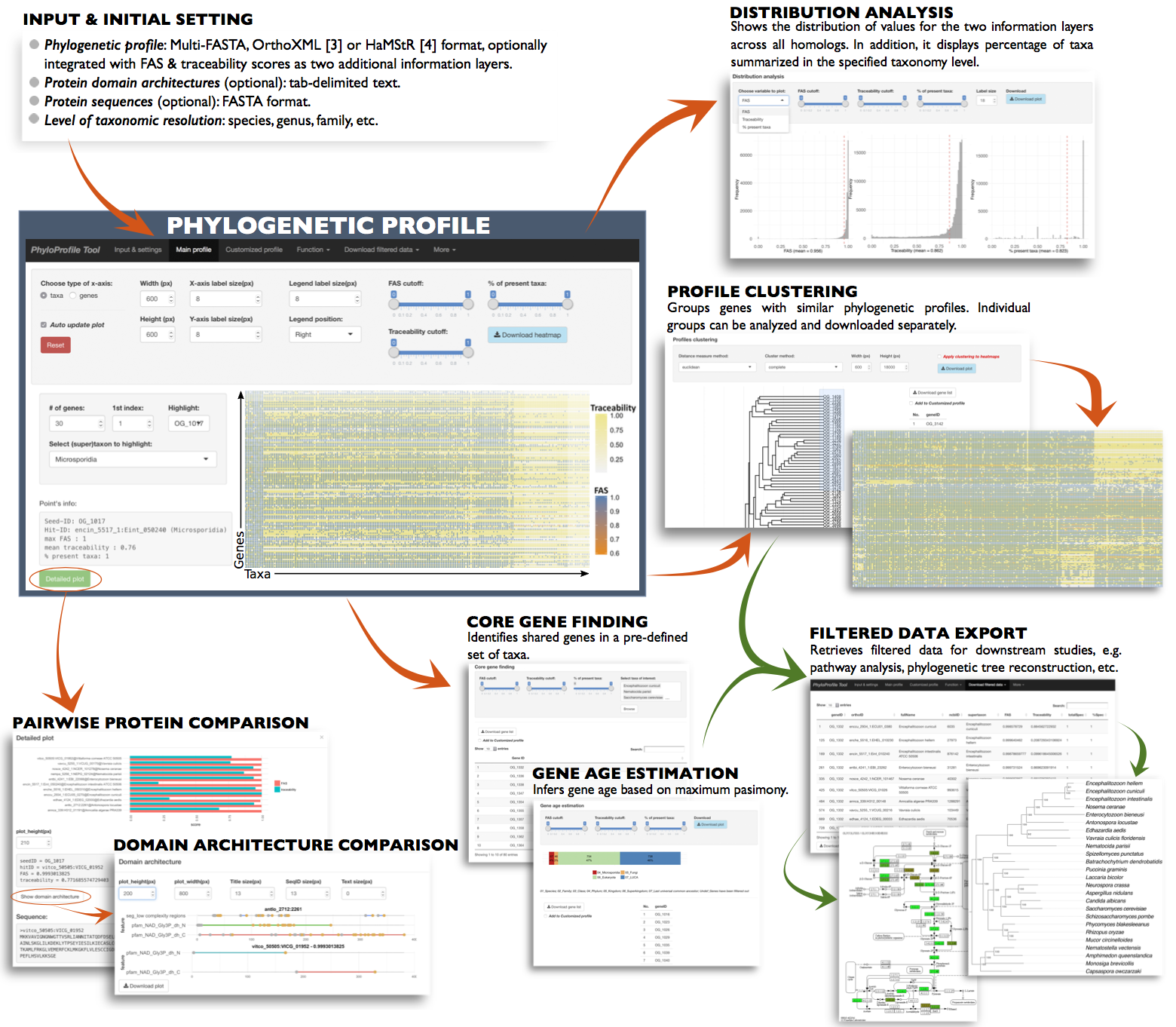
Main input
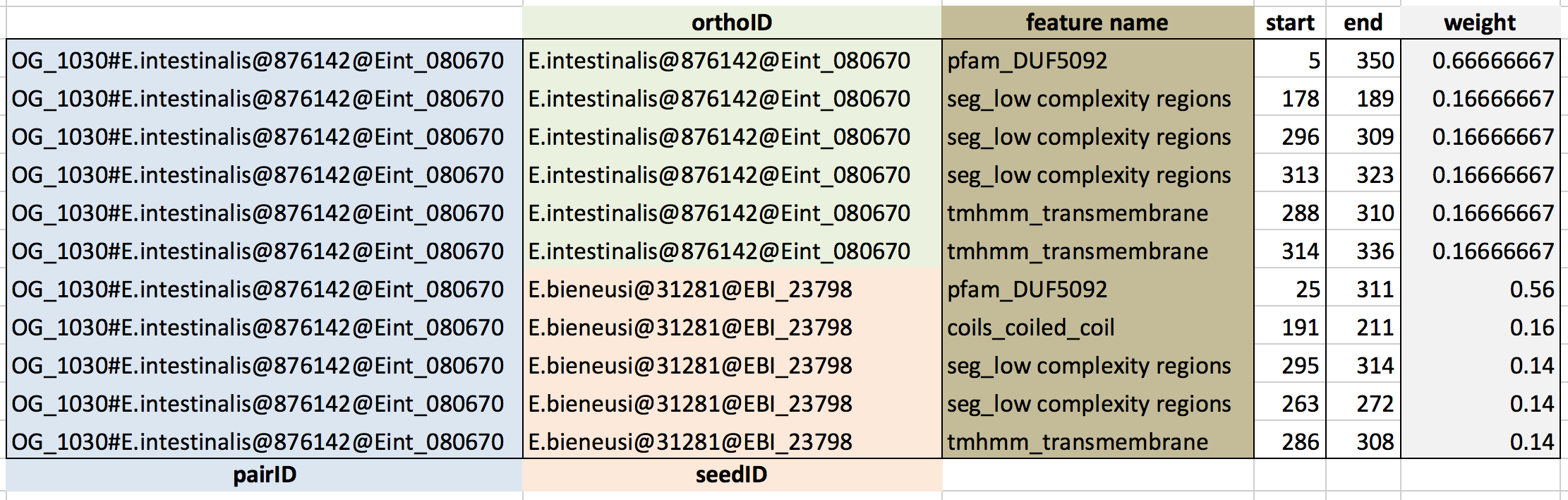
Required information
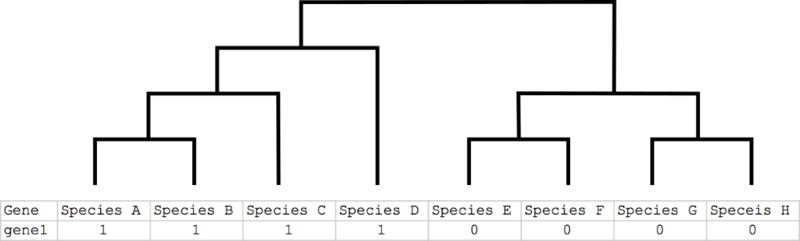
- for basic phylogenetic profile:
- Gene ID of seed protein or ortholog group ID - geneID
- Taxonomy ID of species having orthologs (ncbi+taxonID, e.g. ncbi7029, ncbi3702) - ncbiID (*)
- Ortholog ID - orthoID
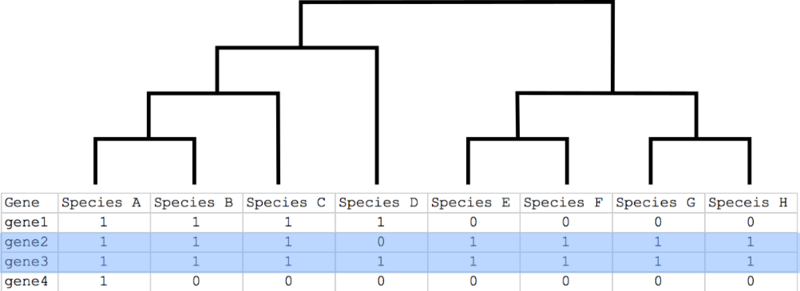
- for additional information layers (optionally):
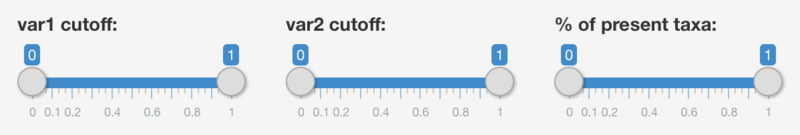
- Value for first additional information layer - var1
- Value for second additional information layer - var2
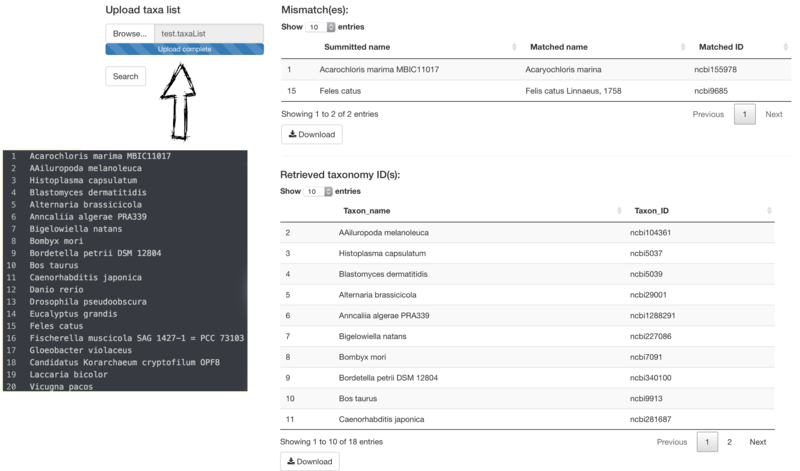
(*) PhyloProfile requires taxonomy IDs in all of input files.
PhyloProfile is provided with a function for searching taxon IDs from a list of taxon names.